Workshop 4 - Testing the gene circuit (11 November)
In our final workshop, we arrive at the final stage of our bacterial photography project based on a study by Tabor et al (2009). Based on their protocol, we inoculated LB media containing ampicillin and streptomycin with E. coli cells of the DH5α strain that had been transformed with the pSR33.4r and the pEDL3 plasmid. The pSR33.4r was the plasmid concerned in the previous Gibson assembly workshops and contains the Cph8, Ho1, and PcyA constructs which are components of the light sensing mechanism. The other plasmid, pEDL3, contained a genetic circuit for edge detection.
On the day we made up agarose consisting of LB media – adding the salt ferric ammonium citrate and S-gal, a substrate used to select recombinant bacterial clones. We then added ampicillin and streptomycin to make sure that only the bacteria that have taken the our plasmids would grow – the two plasmids have genes conferring resistance to these antibiotics. Then we inoculated the agarose with our bacteria and took care to rapidly pour the agarose into plates before they solidify. Although the bacteria are inside the agarose gel, rather than growing as colonies above it, they could still survive as the agarose was not so dense that it prevented air to diffuse in the agarose. We left the plates under the light masks we designed for 24 hours under a red LED lamp. According to the study by Tabor et al, the bacteria is most sensitive to red light.
However, using the DH5α strain of E. coli instead of the JW3367c strain as used by Tabor et al (2009) could result in the salt concentration in the media causing excess downstream signalling from the EnvZ/OmpR system used, as suggested by Foo et al (2015). What this means is that it would be likely that we would see saturation, making the plates all black. Unfortunately, that was exactly what we saw.

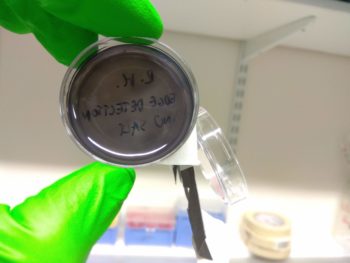
Although it wasn’t quite the edge detection we hoped for, the colour change indicated that the double transformation worked, which is itself a significant achievement!
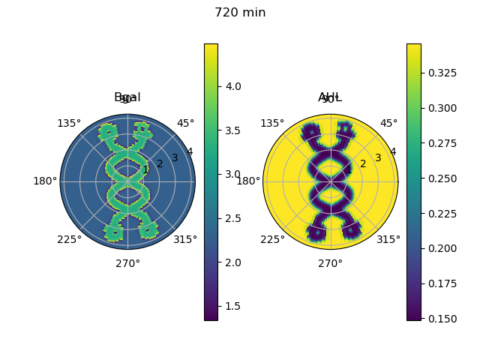
Bill Jia, our president, also went through the Python code he wrote with David Chong, adapting a computational model from the Tabor et al (2009) paper… which could be found here! The script simulated the level of expression of β-galactoside and the signalling molecule AHL at various points in a circular plate placed under a custom light mask over a time course, based on differential equations. So this section of this week’s workshop looks at how computational models can be used to simulate biological processes.
Once again, we thank everyone who has been with us all through this term’s workshops! Our next item on the agenda is planning for our long-term project next term.