Bacterial Edge Detection Modeling
As Lent term comes to a close, the members of the computational subteam of the Bioproject have been wrapping up the coding of the model edge detector system. After some struggles with constants and boundary conditions, we finally succeeded in producing a program that simulates the edge detection circuit!
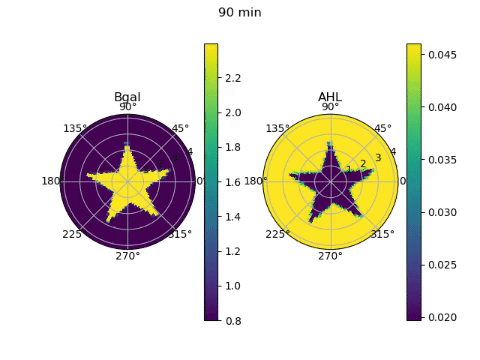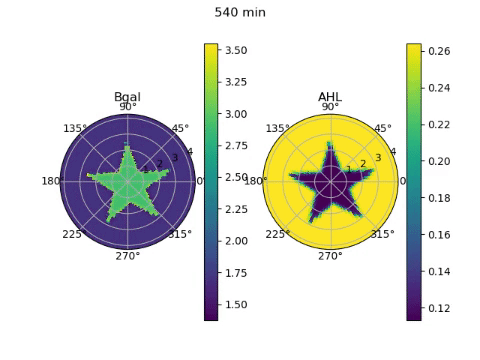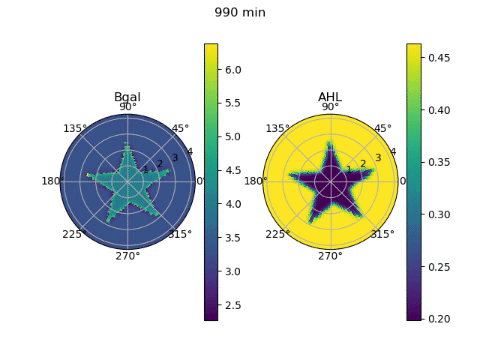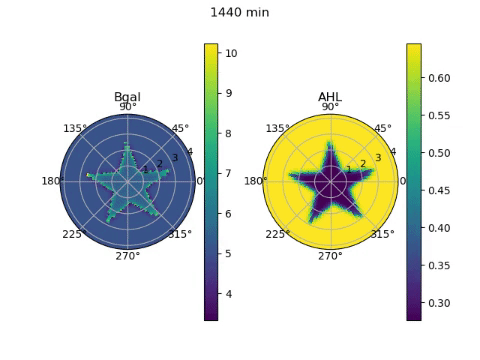
The edge detector works by instructing bacteria under darkness to produce AHL, a diffusible molecule that can turn on gene expression, and CI, a non-diffusable molecule that can turn off gene expression. β-galactosidase, the gene required to ‘ink’ the petri dish with an image, is placed under the control of both these molecules. Since bacteria in darkness produce both molecules, they are unable to produce β-galactosidase due to the effect of the CI. Bacteria exposed to light on the other hand, produce neither molecule and are unable to produce β-galactosidase due to the lack of AHL. On the boundary between darkness and light however, the AHL from light-exposed bacteria can diffuse a short distance into the lit region, effectively giving them AHL without CI. These bacteria can then produce the ink!
To model the behaviour of the edge detector, we had to figure out how the production of AHL and CI responded to light, and how the production of β-galactosidase responded to the concentrations of the two transcription factors. Furthermore, we had to model the diffusion of the AHL. We obtained equations describing these behaviours from the paper that first reported the edge detector (Tabor et al, 2009), and wrote numerical approximations of the derivatives required for the diffusion equation. Looping over time and solving the diffusion equation, we could see the edges slowly appear over 24 hours of culture. But since we simulated it, it only took a couple of seconds on the computer!
Going forward, we are hoping to further explore this model under different conditions to give us new insights, and to develop our own maths that might produce even more complex behaviours. Feel free to use the source code at https://github.com/barrinalo/cusbsedgedetectionmodelling!
Reference
Jeffrey J. Tabor, Howard M. Salis, Zachary Booth Simpson, Aaron A. Chevalier, Anselm Levskaya, Edward M. Marcotte, Christopher A. Voigt, Andrew D. Ellington, A Synthetic Genetic Edge Detection Program, Cell, Volume 137, Issue 7, 2009, Pages 1272-1281, ISSN 0092-8674, https://doi.org/10.1016/j.cell.2009.04.048.