First PCR and Gel at Biomakespace
We rounded off the Michaelmas term with our first practical session at the newly opened Biomakespace! Our aim for the day was to test out the facilities at Biomakespace by running a PCR followed by performing DNA electrophoresis – both very important molecular biology techniques.
PCR, short for Polymerase Chain Reaction, is an extremely useful technique which enables us to massively amplify a specific region of a DNA template molecule. A commonly used template is genomic DNA extracted from tissues or cells, however we were using a selection of BioBricks left over from previous Cambridge iGEM teams. BioBricks are a selection of standardised ‘biological components’ which come inserted into larger circular DNA molecules called plasmids. BioBricks are typically distributed as 2-3 ng dry DNA samples in 384-well plastic plates which we dissolved in water on the day.
Since we plan to use Gibson assembly to build light-sensitive genetic circuits next term, we wanted to selectively isolate the sequence we need from the rest of the plasmid and to customise the ends. To do so, we needed to add carefully selected primers to the PCR samples. Last week (19/11/17) we had a meeting where we discussed some of the basics of good primer selection and designed a few set of primers predicted to amplify the sequences we wanted – thanks to Jarrod Shilts for helping us with this! Primers are small pieces of DNA, around 20 nucleotides long, which can be custom ordered from various companies. Ours came as tiny pellets of dried DNA in tubes which we dissolved and diluted to the working concentration. We had a few sets of primers: one set intended to bind and amplify directly from the start and finish of the coding regions of the BioBrick genes, and another two sets which amplified from common sequences shared between the plasmids our BioBricks were inserted into.
While our PCR was running (on a rather antique thermocycler) we set up the gel we were going to use for separating and visualising the DNA fragments we hoped to generate. DNA electrophoresis relies on the fact that DNA is negatively charged and thus will move from cathode (negative electrode) towards the anode (positive electrode) when placed in an electric field. The gel provides an environment which slows larger pieces of DNA more than smaller ones, which means that smaller ones migrate faster and thus DNA separates by size. By loading a marker sample, with several known-length pieces of DNA, one can estimate the size of DNA in other gel lanes (e.g. our PCR-amplified ones). We had to perform a little DIY to keep the liquid gel in the mould while it set which worked with only a little leakage. Just before pouring we added a dye which fluoresces when in contact with DNA. After running the gel, this dye enabled us to see our PCR products as bright bands when the fluorophore was excited by a blue light transilluminator.
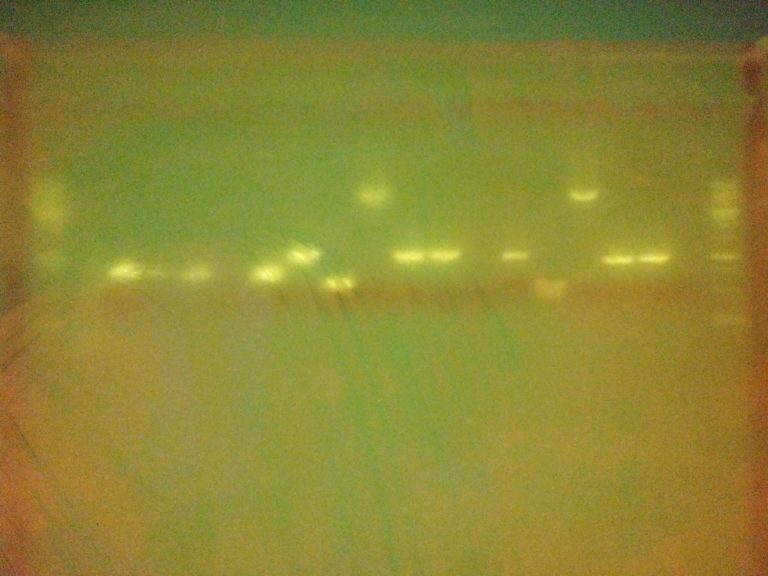
The gel image isn’t as clear as one might like (partly due to a dodgy phone camera!) but there are very clear bands indicating that most of our PCRs were successful! Lane contents: the outer-most lane on each side contained a marker ladder, lanes 2-7 were samples amplified with primers directly over the gene coding regions (designed last week), lanes 8-13 were samples amplified with generic plasmid primers taken from the BioBrick repository website, lanes 14-19 were amplified with similar generic plasmid primers (also designed last week).
Great job to all who helped with the primer design and helped with the practical work! The results indicate that we have several sets of primers which will be useful as start-points for producing overhang primers required to prep sequences for Gibson assembly. There are a few lanes which produced unexpected results though. For example lanes 13 & 19, which used the same template, don’t show bands from either generic primer set. Conversely lanes 10 & 16 showed bands which look a bit larger than we anticipated. These are both things which may need investigating further – something one of our project managers may have time for over the vacation.
Thanks to everyone who has come to our events and meetings this term, we’re expecting things to get more exciting after the winter vacation as we attempt to assemble our light-sensitive circuits and boot them up in E. coli bacteria! Finally, many thanks to Katia Smith-Litière and especially Jenny Molloy for helping us with paperwork, ordering, and more which enabled us to get up and running at Biomakespace this term.