Molecular Biology at Makespace!
Today was our first piece of practical molecular biology for the CUSBS Bioproject 2017-18! We met at the Makespace, Mill Lane, as part of the Open Technology Week Science Make-a-thon. Our goal was to test our ability to produce the enzyme β-galactosidase/LacZ from plasmid DNA in an in vitro gene expression system. This consisted of a commercially available cell-free extract which is essentially the cytoplasm (intracellular fluid) from E. coli bacteria (minus it’s DNA and cellular membranes). The extract still has all the machinery required to create proteins from plasmid DNA templates and there is increasing interest in using this kind of setup for rapidly prototyping synthetic genetic circuits.

We had two plasmids with the gene LacZ under control of a promoter which enables gene expression in the cell-free system. We incubated the plasmids separately in the cell-free solution for around 2 hours at 30°C before adding a coloured indicator of β-galactosidase/LacZ activity. Additionally we included a couple of negative control tubes – these contained only the cell-free extract and were important for verifying whether any colour change we observed could really be attributed to plasmid gene expression. The indicator pigment we were using was Chlorophenol Red-β-D-galactopyranoside (CPRG). This is usually a yellow/orange colour in solution, however β-galactosidase/LacZ can catalyse it’s breakdown into smaller molecules – the result is a shift in colour to red/purple. Shown below:
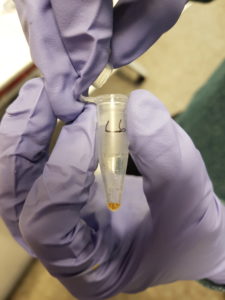
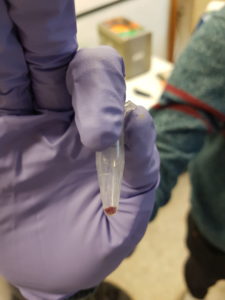
The hope was that in our negative control tubes, ones with no plasmid DNA, we would have seen no colour change since β-galactosidase/LacZ should not have been produced. In the tubes with plasmid we hoped to see a colour change to red/purple, which would indicate that active β-galactosidase/LacZ had been expressed successfully. We did observe a colour change in the tubes with plasmid present. However, we also saw a colour change in our negative control tubes, which indicates that there was likely already β-galactosidase/LacZ present in our cell-free extract. This was a major lesson in how important appropriate controls can be for interpreting scientific data!

While this wasn’t quite the positive result we were hoping to see, it still gives us some useful information; we were considering using cell-free extract to test some of the light-sensing constructs we’re going to produce. The published plasmids we were aiming to use have β-galactosidase/LacZ as their output which we now know would cause us some problems! The plasmids we used today were kindly donated to us by another Cambridge student group, OpenDiagnostics, who are interested in using cell-free extract systems to detect pathogenic viral DNA/RNA. Our data will be important for them to guide the development of suitable reporter mechanisms.
It was fantastic to have so many people interested in student-run synthetic biology; today was just a small taste of the kind of work we’re planning for this year once we move in at Biomakespace and start working on our Bacterial Photography and Image Processing projects. We hope to see you there!